CNIC has made new progress in Influenza Genome Language Model Research
The continuous antigenic variation of influenza viruses is a major cause of recurrent seasonal influenza outbreaks and the frequent need for vaccine updates, posing a long-term challenge to public health efforts. Existing prediction methods based on phylogenetic trees and mutation models struggle to systematically capture the co-evolutionary relationships among different gene segments in the influenza genome, resulting in limited predictive accuracy and generalization ability. Integrating biological structural priors with generative artificial intelligence has therefore become an important research direction in this field.
Recently, the Operation and Application Services Division of the Supercomputing Center, in collaboration with the Beijing Institute of Genomics, Chinese Academy of Sciences (China National Center for Bioinformation), proposed a structure-aware DNA language model, AntigenLM. Built upon an autoregressive Transformer architecture, AntigenLM directly models the complete influenza virus genome during the pretraining stage, explicitly preserving the functional structure and sequential order of the eight gene segments to learn high-order co-evolutionary constraints across segments. During fine-tuning, sentinel markers are introduced to guide the model’s focus toward antigen-related functional regions.
Experimental results demonstrate that AntigenLM reduces the number of amino acid mismatches in HA and NA antigen sequence prediction by approximately 50%–70% compared with existing models, achieving almost zero mismatches in key epitope regions. The model also maintains stable performance in cross-regional transmission scenarios and low-sample subtypes (such as H7N9). In addition, AntigenLM achieves an F1 score of 99.81% in multi-subtype classification tasks for influenza A. This study validates the effectiveness of incorporating biological structural priors as inductive biases into foundation model design.
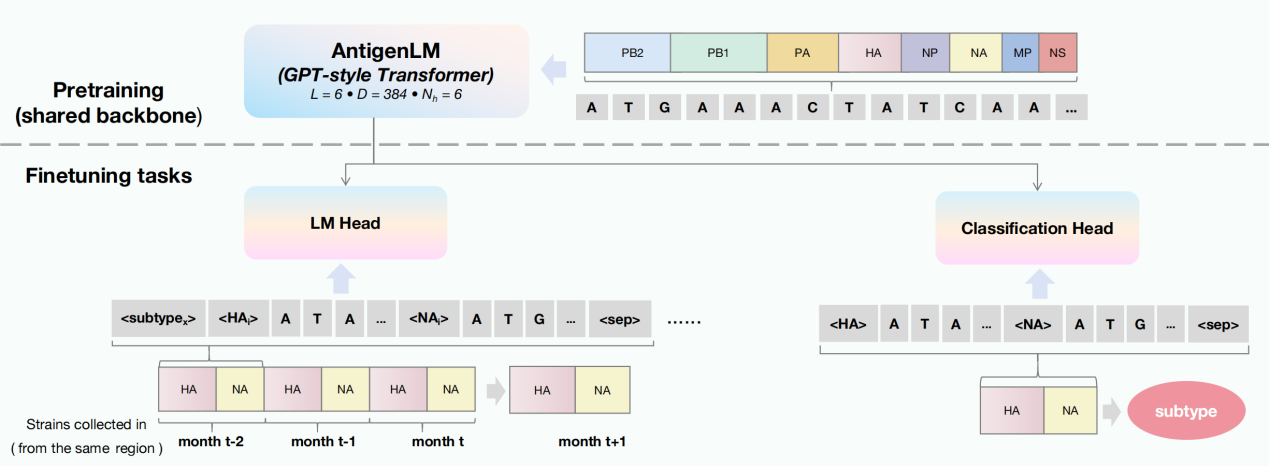
This research has been accepted by the 14th International Conference on Learning Representations (ICLR 2026), one of the top international conferences in machine learning and deep learning. The first author of the paper is Yue Pei, a PhD candidate at our center, and Professor Xuebin Chi is a co-corresponding author. This work was supported by the National Key Research and Development Program of China.
Yue Pei, Xuebin Chi, Yu Kang. AntigenLM: Structure-Aware DNA Language Modeling for Influenza. In The Fourteenth International Conference on Learning Representations (ICLR 2026).
