Research progress on the prediction tool of SUMOylation sites and SUMO-interacting motifs
SUMOylating (SUMO, Small ubiquitin-like modifier), a kind of highly conserved ubiquitination-like modification, plays essential roles in regulating a variety of biological processes, ranging from gene expression and chromatin remodelling to cellular dynamics and plasticity. The dysfunction of SUMOs is closely related to numerous types of diseases, such as neurodegenerative diseases, autoimmune diseases and cancers. Therefore, the identification of SUMOylation sites and SUMO-interacting motifs (SIMs) is of great importance to better understand the roles of SUMOs in cellular, physiological and pathological processes, and facilitate the exploration of potential therapeutic targets for disease treatment.
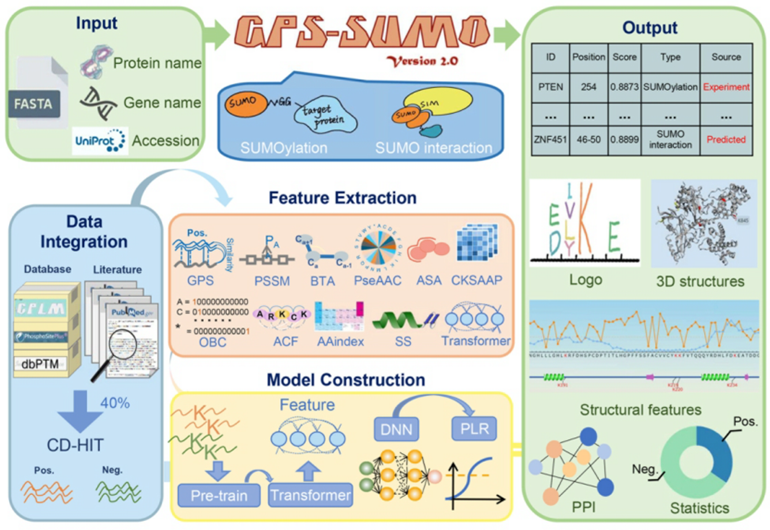
Flowchart of GPS-SUMO 2.0
Collaborated with Prof. Yu Xue 's team from Huazhong University of Science and Technology (HUST), the High Performance Computing Technology and Application Department (HPC Department) of our Center has recently released GPS-SUMO 2.0, a language model for the prediction of SUMOylation sites and SIMs. This transformer-based model was pretrained and fine-tuned with three machine learning algorithms on the ORISE Supercomputer. GPS-SUMO 2.0 not only has demonstrated greater accuracy in predicting SUMOylation sites than other existing tools, but also was capable of annotating the prediction results based on the integrated knowledge from 35 public resources. Therefore, the GPS-SUMO 2.0 was considered to be an effective assistance for experimental screening.
The research results have been published online in Nucleic Acids Research (Impact Factor: 14.9, JCR Q1, CAS journal ranking Q2 TOP). Dr. Teng Lu from HPC Department of our center, and Prof. Yu Xue and Dr. Di Peng from School of Life Science and Technology, HUST are co-corresponding authors. This work was supported by the National Key R&D Program of China, the National Natural Science Foundation of China and the Strategic Priority Research Program of CAS.
